Athenomics - Consideration of NGS library prep for FFPE samples
Performing mRNA-seq on FFPE samples is feasible, but presents unique challenges due to degraded and chemically modified RNA
NGS Sample Types and Considerations for Library Preparation
NGS libraries can be generated from a wide variety of sample types, including fresh-frozen tissue biopsies, blood specimens, formalin-fixed paraffin-embedded (FFPE) tissues, fine-needle aspirates (FNAs), core-needle biopsies (CNBs), liquid biopsies, and even isolated single cells. However, not all samples are equally suitable for every NGS method; certain techniques require larger tissue amounts or high-quality nucleic acids, and may not perform optimally with degraded material that is common in clinical or archival specimens. Therefore, it is essential to choose a library preparation method that is well-matched to both the sample’s characteristics and the specific research objectives.
Reliability of NGS results relies heavily on variables such as nucleic acid quantity, molecular integrity, processing workflows, storage conditions, and, for tumor samples, the percentage of tumor cells (tumor purity). High-quality, high-molecular-weight DNA or RNA from fresh-frozen tissues or blood is generally optimal for most NGS applications. In contrast, degraded or fragmented nucleic acids—often encountered in FFPE tissues, FNAs, and liquid biopsies—necessitate specialized approaches. For these challenging sample types, targeted sequencing methods are favored because they require less input material and are more resilient to damage-induced artifacts.
Working with FFPE Samples
Formalin-fixed paraffin-embedded (FFPE) tissue is the gold standard for clinical and research tissue preservation, as well as the most frequently analyzed sample type in NGS workflows. The preservation process involves thin tissue sectioning and formaldehyde fixation, which stabilizes the cellular architecture but compromises nucleic acid integrity. Formaldehyde fixation induces protein-DNA crosslinks, fragmentation, and the formation of sequencing artifacts, notably cytosine-to-thymine transitions via deamination. As a result, FFPE-derived DNA is highly fragmented (typically <300 bp) and the overall quality depends on the fixation protocol, duration of storage, and cellular composition.
Performing mRNA-seq on FFPE samples is feasible, but presents unique challenges due to degraded and chemically modified RNA. Both RNA extraction and library preparation must be carefully optimized for FFPE inputs. Random-primed cDNA synthesis is particularly effective for transcriptome studies from FFPE and other fragmented RNA samples, but ribosomal RNA—which can make up more than 90% of total RNA—must be removed prior to cDNA synthesis to ensure a high-quality transcriptomic analysis.
It is a common concern that the 3’ mRNA sequencing method may not be well suited for FFPE samples because of these issues. However, several studies have demonstrated that 3′ mRNA sequencing reliably generates gene expression profiles that are comparable to those produced by whole transcriptome sequencing methods, showing that both approaches are effective and suitable for analyzing archived FFPE samples.
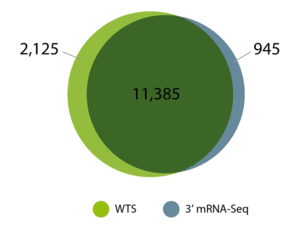
Each sample type for NGS analysis presents unique challenges and considerations. Our team has extensive experience with FFPE tissue and other difficult sample types, and can provide guidance and support to optimize your NGS workflows.
Source of figure: Transcriptomics on FFPE Samples: 3’ mRNA-Seq vs. Whole Transcriptome Sequencing