Athenomics - How to choose the right sequencing technology
Choosing the right sequencing platform depends on factors like project goals, budget, accuracy requirements, and data output.
Introduction
DNA sequencing has revolutionized biological research, medicine, and biotechnology. With rapid advancements in sequencing technologies, researchers now have multiple options, each with unique strengths and limitations. Choosing the right sequencing platform depends on factors like project goals, budget, accuracy requirements, and data output.
In this blog, we’ll explore the major sequencing technologies available today, compare their key features, and provide a step-by-step guide to help you select the best option for your needs.
1. Understanding Different Sequencing Technologies
Before selecting a sequencing platform, it’s essential to understand the different technologies available. Here’s an overview of the most widely used methods:
A. Sanger Sequencing (First-Generation Sequencing)
- Principle: Chain-termination method using fluorescently labeled dideoxynucleotides.
- Read Length: Up to 1,000 bp.
- Throughput: Low (single reactions or small batches).
- Best For: Small-scale projects, validation, cloning, and targeted sequencing.
B. Next-Generation Sequencing (NGS) (Second-Generation Sequencing)
NGS platforms enable massively parallel sequencing, making them ideal for large-scale genomic studies. The most common NGS technologies include:
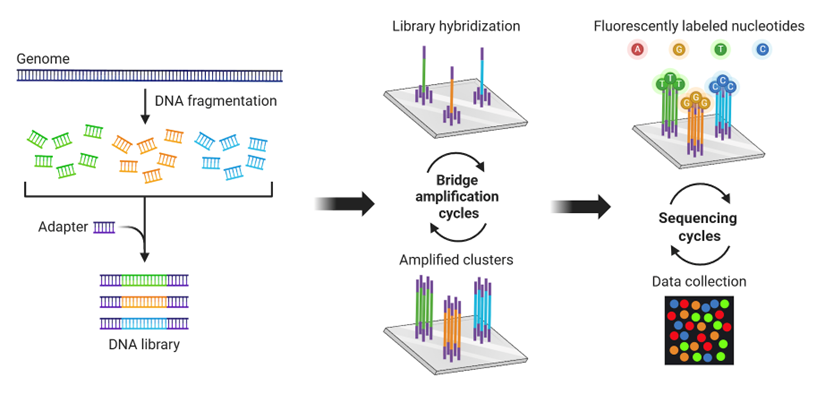
i. Illumina (Short-Read Sequencing)
- Principle: Sequencing-by-synthesis with reversible terminators.
- Read Length: 50-300 bp (pair-end).
- Throughput: Very high (up to terabytes of data per run).
- Best For: Whole-genome sequencing (WGS), exome sequencing, RNA-seq, ChIP-seq.
ii. Ion Torrent (Semiconductor Sequencing)
- Principle: Detects hydrogen ions released during DNA polymerization.
- Read Length: Up to 400 bp.
- Throughput: Moderate (faster run times than Illumina).
- Best For: Targeted sequencing, microbial genomics, clinical diagnostics.
C. Third-Generation Sequencing (Long-Read Sequencing)
These technologies generate much longer reads, overcoming limitations of short-read sequencing.
i. PacBio (SMRT Sequencing)
- Principle: Real-time sequencing using fluorescent nucleotides in zero-mode waveguides.
- Read Length: 10,000–100,000 bp.
- Throughput: Moderate.
- Best For: De novo genome assembly, structural variant detection, full-length transcriptomics.
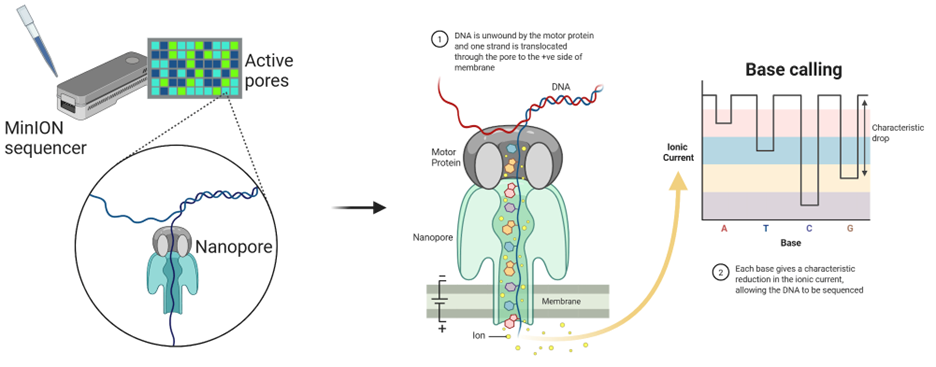
ii. Oxford Nanopore (Nanopore Sequencing)
- Principle: Measures changes in electrical current as DNA passes through a nanopore.
- Read Length: Up to 2 Mb (theoretical limit).
- Throughput: Flexible (MinION for portable use, PromethION for high throughput).
- Best For: Real-time sequencing, field applications, metagenomics, large structural studies.
2. Key Factors to Consider When Choosing a Sequencing Technology
A. Project Goals
- Whole-genome sequencing (WGS): Illumina (cost-effective), PacBio/Nanopore (for complex genomes).
- Targeted sequencing (e.g., exomes, panels): Illumina or Ion Torrent.
- Metagenomics: Nanopore (real-time) or Illumina (high accuracy).
- Structural variant detection: PacBio or Nanopore (long reads).
- Clinical diagnostics: Illumina (accuracy) or Ion Torrent (speed).
B. Read Length Requirements
- Short-read (Illumina/Ion Torrent): Sufficient for most applications.
- Long-read (PacBio/Nanopore): Needed for repetitive regions, structural variants, or de novo assembly.
C. Accuracy Needs
- Highest accuracy (~99.9%): Illumina or Sanger.
- Moderate accuracy (~95%): Nanopore or PacBio (can be improved with consensus sequencing).
D. Budget and Cost-Effectiveness
- Low-budget projects: Sanger (small scale) or Illumina (large scale).
- High-budget, long-read projects: PacBio or Nanopore.
E. Turnaround Time
- Fast results (hours): Nanopore (real-time).
- Standard (days-weeks): Illumina, PacBio.
F. Sample Type and Quality
- Degraded DNA (e.g., FFPE samples): Short-read sequencing (Illumina).
- High-molecular-weight DNA: Long-read sequencing (PacBio/Nanopore).
3. Decision-Making Workflow
- Define your research question: What do you need to sequence, and why?
- Assess sample type and quality: Will your DNA/RNA work with the chosen technology?
- Evaluate read length and accuracy needs: Do you need long reads or high precision?
- Consider throughput and scalability: How many samples will you process?
- Check budget constraints: Can you afford the instrument, reagents, and analysis?
- Review bioinformatics requirements: Do you have the computational resources?
4. What’s New in 2025?
Sequencing is a rapidly expanding field. As of mid-2025, several new sequencing platforms and updates to existing technologies have been launched, pushing the boundaries of speed, accuracy, cost-efficiency, and portability. Here’s a roundup of the latest advancements in DNA/RNA sequencing technologies:
- Roche Sequencing by Expansion (SBX): Announced in early 2025, SBX introduces a novel chemistry that amplifies DNA into “Xpandomers,” allowing rapid and accurate base-calling with CMOS-based detection. It’s ideal for high-throughput genome sequencing with a short turnaround. SBX will launch in 2026.
- Illumina’s 5-Base Chemistry: This new capability enables detection of standard bases and methylation states in a single run. Expected to revolutionize epigenetic and variant analysis in multi-omic workflows.
- Element Biosciences’ AVITI Prime: The AVITI sequencer provides Q40-level accuracy (99.99%) and 300 bp reads with new polymerase chemistry. It features flexible throughput up to 1B reads per run. This sequencer is best for single-cell sequencing, cancer genomics, and precision medicine.
- MGI’s DNBSEQ-T20 (2025 Launch – “The Hyper-Sequencer”): DNBSEQ-T20 offers massive throughput up to 50 Tb per run (designed for million-genome projects), combining high speed and accuracy at an affordable cost. It is best suited for national-scale genomics initiatives, biobanks, and industrial-scale sequencing.
Conclusion
Choosing the right sequencing technology depends on balancing accuracy, read length, cost, and project goals. By carefully evaluating your needs and comparing different platforms, you can select the best sequencing method to achieve reliable and meaningful results. Contact us for a free consultation about your projects.