Athenomics - Introduction of Next-Generation Sequencing (NGS) Technology
The breakthrough of NGS has transformed fields such as genetic research, precision medicine, cancer genomics, infectious disease surveillance, and agriculture.
Next-Generation Sequencing (NGS) has revolutionized genomics by enabling rapid, cost-effective, and high-throughput DNA and RNA sequencing. Unlike traditional Sanger sequencing, which processes one DNA fragment at a time, NGS allows millions to billions of DNA strands to be sequenced simultaneously. This breakthrough has transformed fields such as genetic research, precision medicine, cancer genomics, infectious disease surveillance, and agriculture.
A typical NGS experiment shares similar steps regardless of the instrument technology used.
Step 1: DNA/RNA Extraction
High-quality nucleic acids are extracted from samples, which may include blood, tissue, saliva, and other biological materials. If RNA is to be sequenced, it must first be converted into complementary DNA (cDNA).
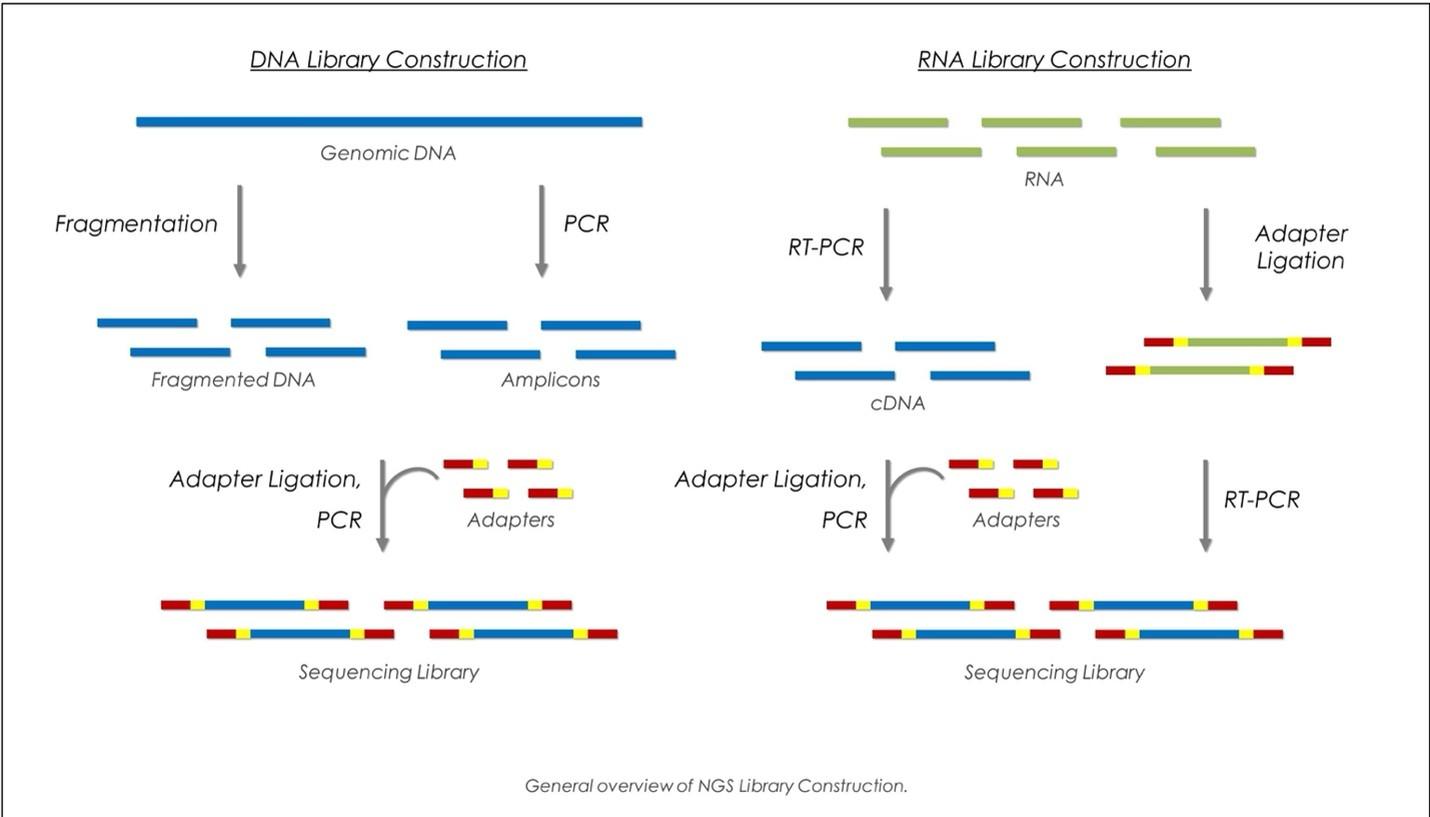
Step 2: Library Preparation
Preparing DNA or RNA for sequencing involves several essential substeps:
Fragmentation
DNA is sheared into small fragments, typically ranging from 200 to 800 base pairs. This can be achieved through mechanical shearing methods such as sonication or nebulization, or via enzymatic fragmentation using transposase-based technologies.
Adapter Ligation
Short DNA adapters are ligated to the ends of the DNA fragments. These adapters facilitate primer binding during sequencing and allow for barcoding, enabling the multiplexing of multiple samples within a single run.
Amplification (Optional)
Polymerase Chain Reaction (PCR) may be employed to enrich library fragments and increase input material for sequencing—note that some single-molecule platforms (such as PacBio and Nanopore) do not require amplification.
Quality Control
Final library products are quality-checked for fragment size and concentration, using instruments such as the Bioanalyzer or Qubit.
Step 3: Sequencing
Prepared libraries are loaded onto sequencing instruments such as those manufactured by Illumina or Nanopore. Each sequencing platform utilizes specific detection methods:
- Illumina: Detects fluorescent signals from reversible terminators.
- Ion Torrent: Identifies pH changes during nucleotide incorporation.
- Nanopore: Measures changes in electrical current as DNA passes through nanopores.
Step 4: Data Analysis
Once sequencing is complete, raw data (usually in FASTQ format) undergoes several analysis steps:
- Base Calling
- Quality Control (QC)
- Read Alignment
- Variant Calling and Annotation
- Downstream Analysis
Conclusion
NGS is a powerful, versatile, and rapidly evolving technology that has transformed genomics. Each step, from library preparation to data analysis, is critical for generating accurate insights. As costs decrease and accuracy improves, NGS will continue to drive innovation in medicine, agriculture, and biotechnology.
_For more information or to apply NGS in your research, consult with sequencing specialists or bioinformaticians to help design your experiment.
Source of figure: biocompare.com: Next-Generation Sequencing