Athenomics - RNA-seq library prep by poly (A) tailing
Poly(A) tailing can be used for library preparation of RNAs lacking a natural poly(A) tail, such as miRNA and other small RNAs. This blog discusses the application of poly(A) tailing in RNA-seq.
Despite the routine use of traditional RNA-seq in transcriptomics, standard RNA library preparation methods often require good-quality RNA with a RIN ≥ 8 and tend to offer limited sample multiplexing capabilities. These limitations present significant challenges for large-scale studies that depend on poor-quality RNA obtained from archival tissues stored in biobanks and preserved using fixation methods that are highly destructive to RNA.
To unlock transcriptomic discoveries hidden within degraded RNA from these valuable repositories, researchers have incorporated polyadenylation of degraded total RNA fragments into RNA-seq library preparation. Poly(A) tailing of RNA enables the generation of robust, reliable, and reproducible full-length transcriptome data from even the poorest quality RNA, including samples fixed by any method. Additionally, poly(A) tailing can be used for library preparation of RNAs lacking a natural poly(A) tail, such as miRNA and other small RNAs. This blog discusses the application of poly(A) tailing in RNA-seq.
Poly(A) Tailing for Small RNA Library Preparation
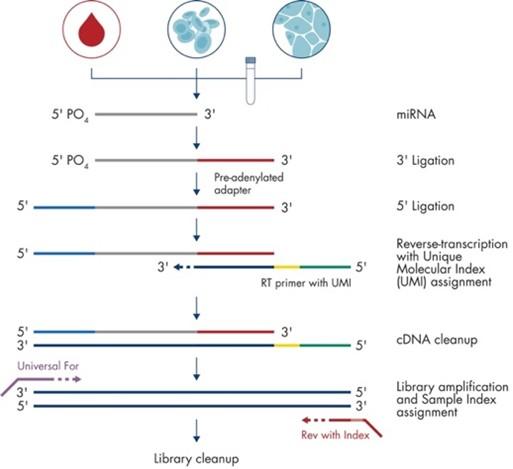
Conventional small RNA sequencing library preparation involves sequential 3' and 5' adapter ligation, followed by reverse transcription and PCR amplification. However, ligation-based approaches often suffer from low sensitivity and substantial bias, compromising the accurate representation of cellular small RNA populations. To overcome these limitations, the poly(A) tailing technique was developed as an alternative strategy. This approach employs poly(A) polymerase to efficiently add poly(A) tails, outperforming traditional ligation methods in both efficiency and reliability. In this method, the 3' adapter is incorporated during reverse transcription via an oligo-dT primer, while the 5' adapter is added through template switching. Compared with standard ligation-based protocols, the poly(A) tailing approach offers advantages in cost-effectiveness and reduced processing time.
Poly(A) Tailing for Full-Length mRNA-seq
The standard workflow for full-length mRNA-seq library preparation includes multiple steps such as RNA fragmentation, reverse transcription, adapter ligation, and PCR amplification. This process is inherently complex, requiring precise optimization and purification to maintain high library quality. Commercial kits for full-length mRNA library construction are typically more expensive than targeted RNA-seq methods (for example, 3' end counting), which can limit their use in large-scale projects. The technical challenges and increased expenses are mainly due to the need for complete transcript representation and the minimization of biases during library construction.
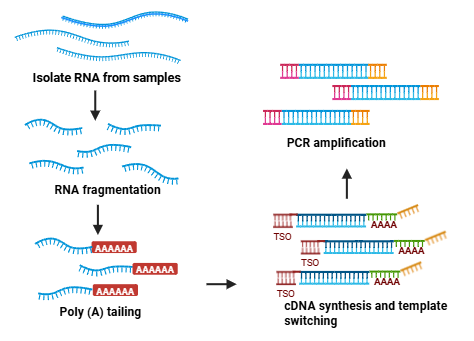
RNA isolated from FFPE (formalin-fixed, paraffin-embedded) specimens is often degraded and fragmented, posing major challenges for conventional RNA-seq workflows. Low RNA quality degrades the reliability of transcriptome profiling with standard approaches. Highly degraded mRNA frequently lacks an intact poly(A) tail, limiting the usefulness of 3'-focused mRNA-seq methods.
The poly(A) tailing strategy provides a cost-effective and streamlined alternative for full-length mRNA-seq library preparation and performs reliably with both high-quality and degraded RNA. Building on this, the recently developed Strand-optimized Smart-seq (So-Smart-seq) achieves similar efficiency with polyadenylated RNAs but also improves the detection of non-polyadenylated transcripts. Furthermore, the MERCURIUS™ FFPE-seq method is specifically optimized for severely degraded RNA from FFPE or chemically fixed samples, providing superior full-transcript coverage even in challenging conditions. These innovations collectively expand full-length mRNA sequencing to a broader range of RNA quality scenarios.
Source of figure: QIAseq miRNA Library Kit