Athenomics - Single-Cell RNA Sequencing vs. Single-Nucleus RNA
Single-cell RNA sequencing and single-nucleus RNA sequencing share similar underlying principles, each method offers distinct advantages that suit different research applications
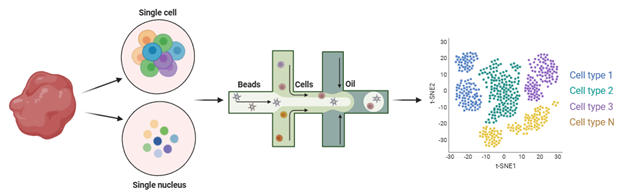
Single-cell RNA sequencing (scRNA-seq) and single-nucleus RNA sequencing (snRNA-seq) are powerful approaches for analyzing transcriptomic heterogeneity at the individual cell level. While they share similar underlying principles, each method offers distinct advantages that suit different research applications. Here, we review the primary considerations for selecting between these techniques and provide an overview of their respective uses in scientific studies.
scRNA-seq
scRNA-seq remains the leading method for investigating cellular heterogeneity, supported by a wide range of established technologies and workflows. Its main advantage is that it captures both cytoplasmic and nuclear transcripts, providing a more complete transcriptomic profile. scRNA-seq typically achieves higher throughput compared to snRNA-seq, making it more scalable for large single-cell studies. It is especially suited for freshly dissociated tissues or cells in suspension.
However, scRNA-seq does have important limitations. Some tissue types require extensive processing or may be incompatible with single-cell isolation. The dissociation process can be labor-intensive and potentially introduce cellular stress or bias, sometimes leading to disproportionate capture of certain cell types, such as immune cells. This may result in skewed data, particularly when analyzing tissues that are difficult to dissociate.
snRNA-seq
snRNA-seq is a high-throughput technique for profiling transcriptomes across thousands of cells by sequencing RNA from isolated nuclei. Its greatest advantage is compatibility with a wide variety of samples, including fresh, frozen, and chemically preserved tissues. snRNA-seq excels with tissue types that are challenging to dissociate while maintaining high quality data. It is especially effective for studying complex cells such as kidney cells, cardiomyocytes, and neurons.
snRNA-seq minimizes dissociation-induced transcriptional stress, a common artifact in scRNA-seq, and consistently provides more biologically relevant data. It is particularly valuable for profiling rare or hard-to-isolate cell populations—such as in studies of tumor heterogeneity—where scRNA-seq often struggles. However, snRNA-seq only analyzes nuclear RNA; it excludes cytoplasmic transcripts. This restriction may limit the detection of RNAs with low nuclear abundance and reduce applicability for some research questions.
What are the Differences between scRNA-seq and snRNA-seq?
The major distinction between scRNA-seq and snRNA-seq is their approach to sample preparation and the source of RNA. scRNA-seq isolates intact single cells, enabling comprehensive profiling of both cytoplasmic and nuclear RNA. snRNA-seq extracts nuclei alone, focusing exclusively on nuclear RNA.
It is reported that both approaches yield comparable overall gene detection rates, but each captures slightly different aspects of the transcriptome, making them complementary in certain applications.
Key Differences Summarized
| Feature | scRNA-seq | snRNA-seq |
|---|---|---|
| Cell Source | Whole cells | Isolated nuclei |
| Tissue Type | Fresh or freshly frozen | Fresh, frozen, or preserved |
| RNA Capture | Total cellular RNA | Nuclear RNA |
| Cell Integrity | Higher risk of damage | More gentle on cells |
| Gene Length Bias | Less pronounced | More pronounced |
| Data Complexity | High (whole transcriptome) | High (nuclear-focused) |
Considerations
Tissue dissociation presents technical challenges for both methods, but is generally more demanding for scRNA-seq because it requires intact single-cell suspensions and careful reagent optimization. Both methods yield similar rates of cell recovery and gene detection, yet comparative studies show each preferentially detects distinct cell populations from the same sample. Rather than complete overlap, the datasets are often complementary.
Choosing between scRNA-seq and snRNA-seq requires careful evaluation of their unique strengths. scRNA-seq is preferred for studies focusing on stress-resistant, easy-to-dissociate cells, and when comprehensive profiling of all transcripts—including cytoplasmic—is required. snRNA-seq is better suited to resolving difficult tissues or archived samples, minimizing cellular stress, and preserving fragile or rare cell types. Practical factors such as sample availability and handling should also influence the decision. Ultimately, the optimal choice depends on the samples and the specific scientific objectives, as both techniques provide distinct biological insights.