Athenomics - What Is Shotgun Metagenomic Sequencing
Shotgun metagenomic sequencing provides a comprehensive, unbiased approach to microbial community analysis by sequencing all DNA content within a sample.
Shotgun metagenomic sequencing provides a comprehensive, unbiased approach to microbial community analysis by sequencing all DNA content within a sample. This high-throughput method differs fundamentally from amplicon-based techniques by avoiding primer-specific amplification, instead employing random fragmentation and sequencing of total extracted DNA. Consequently, it simultaneously captures genomic material from all microbial domains—including bacteria, viruses, fungi, and archaea—enabling a complete taxonomic and functional profile of the microbiome.
Shotgun metagenomics delivers a comprehensive view of microbial ecosystems by simultaneously characterizing taxonomic composition, functional gene content, and metabolic potential. This approach proves particularly valuable for analyzing intricate microbial communities in diverse environments—from soil and aquatic systems to human gut and skin microbiomes. A key advantage lies in its ability to uncover novel microorganisms and rare functional elements that often evade detection through targeted sequencing methods.
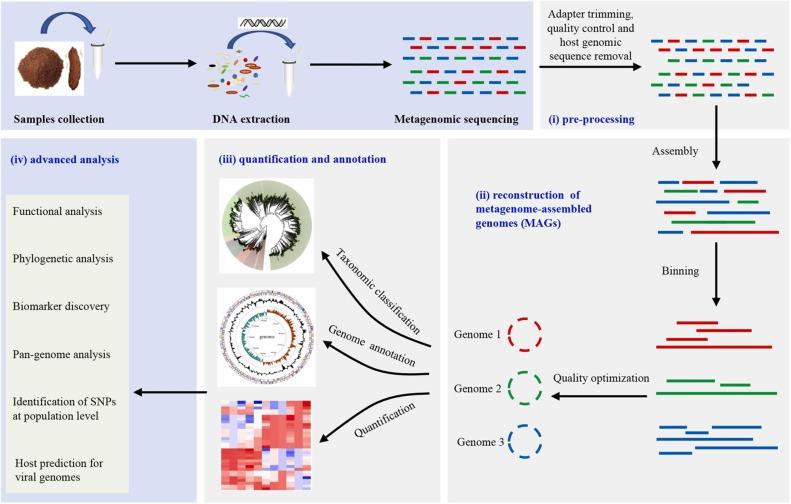
Why Use Shotgun Metagenomic Sequencing?
Shotgun sequencing delivers a comprehensive view of the microbiome—ideal for applications that demand depth, resolution, and functional clarity.
Key Advantages of Shotgun Metagenomics
• Unbiased Profiling: Eliminates primer bias by capturing complete microbial genomes without requiring prior sequence knowledge.
• Strain-Level Precision: Delivers exceptional taxonomic resolution for differentiating closely related microbial strains.
• Functional Characterization: Reveals metabolic pathways, gene functions, and biological mechanisms within microbial communities.
• Comprehensive Detection: Identifies diverse organisms—including viruses, fungi, and previously unculturable species—in a single analysis.
• Discovery Potential: Facilitates de novo genome assembly and identification of novel genetic elements.
• Multi-Omics Ready: Generates data-rich outputs compatible with advanced bioinformatics pipelines and systems biology integration.
Shotgun Metagenomics vs. Amplicon Sequencing
| Feature | Shotgun Metagenomics | Amplicon Sequencing (16S/ITS) |
|---|---|---|
| Approach | Untargeted, sequences all DNA | Targeted, amplifies specific gene regions |
| Coverage | Whole microbial genomes | Only hypervariable regions (e.g., V3-V4) |
| Taxonomic Resolution | Strain-level identification | Typically genus/species level |
| Functional Data | Yes (genes, pathways, AMR markers) | No |
| Organisms Detected | Bacteria, viruses, fungi, archaea | Bacteria (16S) or fungi/yeasts (ITS) only |
| Primer Bias | None | High (affected by primer choice) |
| Cost | Higher ($/sample) | Lower ($/sample) |
| Compute Needs | High (complex bioinformatics) | Moderate |
| Best For | Functional potential, novel discoveries | Taxonomic profiling, large sample numbers |
When to Choose Which
• Shotgun: When you need functional insights, detect viruses/fungi, or require strain-level detail
• Amplicon: For cost-effective taxonomy studies or when processing thousands of samples
Our collaborators have a complete set of devices to accelerate the discovery of microbes. Together with our established shotgun and 16S/18S/ITS sequencing platform, we are able to characterize and isolate microbes in a cost-efficient manner.
Source of figure: DOI: 10.1016/j.micres.2022.127023